scNym: Semi-supervised adversarial neural networks for single cell classification
- 4 minsscNym Paper (Open Access): https://doi.org/10.1101/gr.268581.120
Research Website: scnym.research.calicolabs.com
Single cell genomics allows for simultaneous molecular profiling of thousands of diverse cells and has advanced our understanding of development, aging, and disease. To derive biological insight from these data, each single cell molecular profile must be annotated with a cell identity, such as a cell type or state label. Traditionally, this task has been performed manually by domain expert biologists. Manual annotation is time consuming, somewhat subjective, and error prone, motivating algorithmic approaches.
Many existing single cell classification tools use information in a labeled training set to learn a cell classification model. We usually also have access to an unlabeled “target” dataset that we’d like to label. This target data contains information about the myriad sources of biological and technical bias and batch effects that may have influenced these measurements. Semi-supervised learning methods have demonstrated that using information in the target data during model training can improve performance, but not existing methods take advantage of this extra information.
To make use of information in the unlabeled target dataset for cell type classification, we have developed a semi-supervised, adversarial neural network model scNym.
Semi-supervision and adversarial training
In the typical supervised learning framework, the model touches the target unlabeled dataset to predict labels only after training has concluded. In contrast, our semi-supervised adversarial learning framework trains the model parameters on both the labeled and unlabeled data. scNym uses the unlabeled target data through a combination of MixMatch semi-supervision (Berthelot et. al. 2019) and by training a domain adversary (Ganin et. al. 2016).
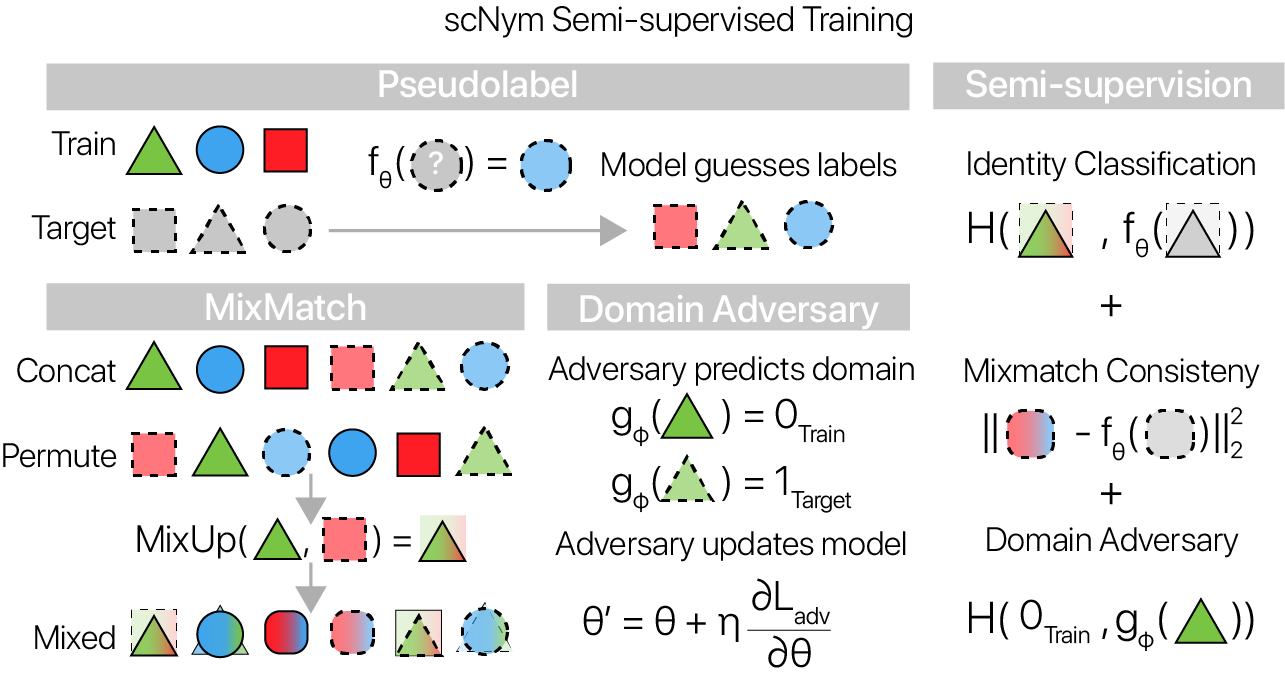
MixMatch
In the MixMatch semi-supervision framework, unlabeled cell profiles from the target dataset are “pseudolabeled” during training using the current classification model. The entropy of these pseudolabels is minimized with a scaling method. This entropy minimization enforces the assumption that each cell belongs to a single class.
After pseudolabeling, the training and target datasets are passed through a randomized “MixUp” operation (Zhang et. al. 2017). Stately simply, both train and target observations are randomly paired, and weighted averages of each pair are computed. We train scNym models to minimize a supervised classification loss on “mixed” training examples and to minimize an interpolation consistency loss on “mixed” target examples.
Domain adversarial training
We also introduce a “domain adversary” during scNym model training. The domain adversary is a simple 2-layer neural network that tries to classify the domain of origin (target or training) for each example given an embedding vector from the scNym classifier. If the adversary performs well, the domains are fairly separated in the embedding, whereas if the adversary performs poorly, the domains are “adapted” and well-mixed in the embedding.
We use the “gradient reversal” trick to allow the classification model to compete with the adversary, helping to adapt across domains in the classifier embedding. At each backward pass, the domain adversary tries to minimize the adversarial classification loss while the classification model tries to maximize it. We found that this training scheme allows scNym models to adapt across training and target domains despite systematic biological and technical differences.
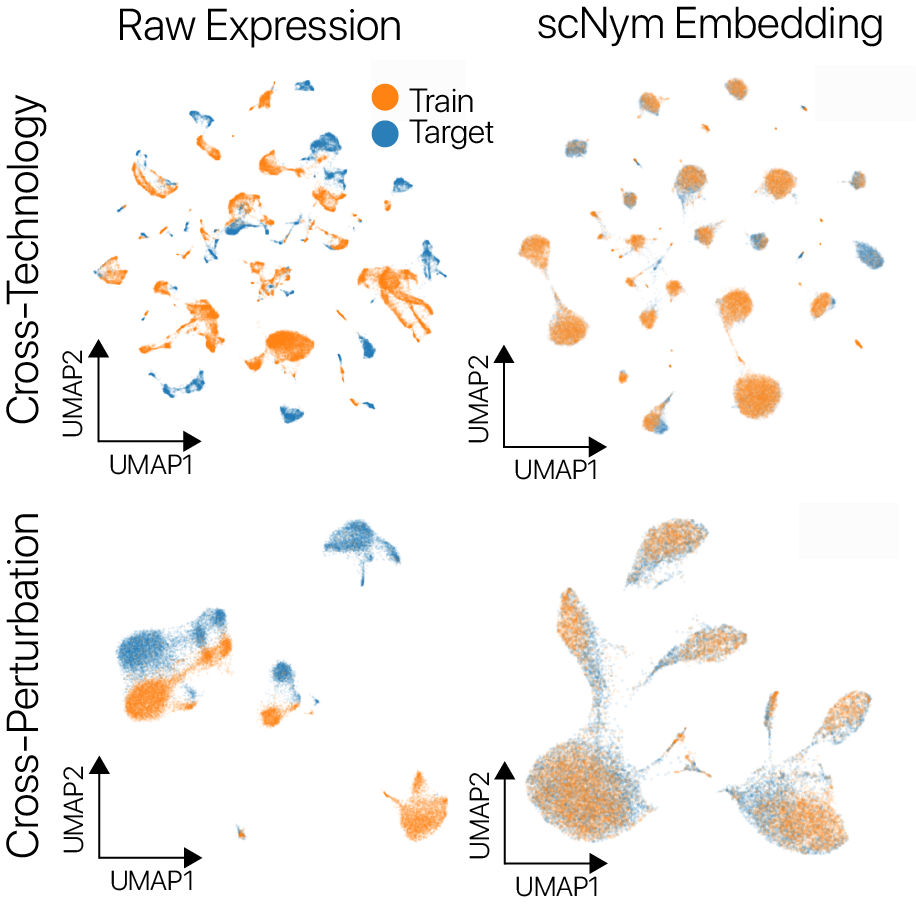
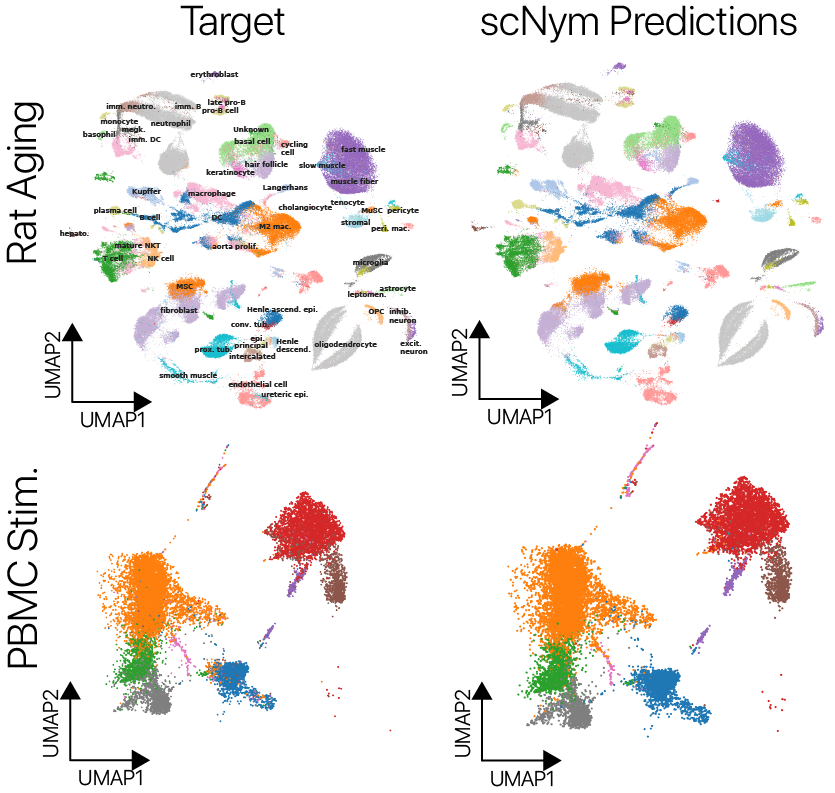
scNym models offer superior single cell classification performance
We trained scNym models to transfer labels across experiments with several different types of systematic variation. We first transferred labels from young rat cells to aged rat cells and found superior performance to existing methods.
We also transferred labels across perturbation conditions, training on unstimulated human peripheral blood mononuclear cells and predicting on type I interferon stimulated cells. We again found superior performance to baseline methods. On several cross-technology benchmarks, we likewise found superior performance to existing methods, despite large differences in sequencing technologies and large class imbalances across data sets.
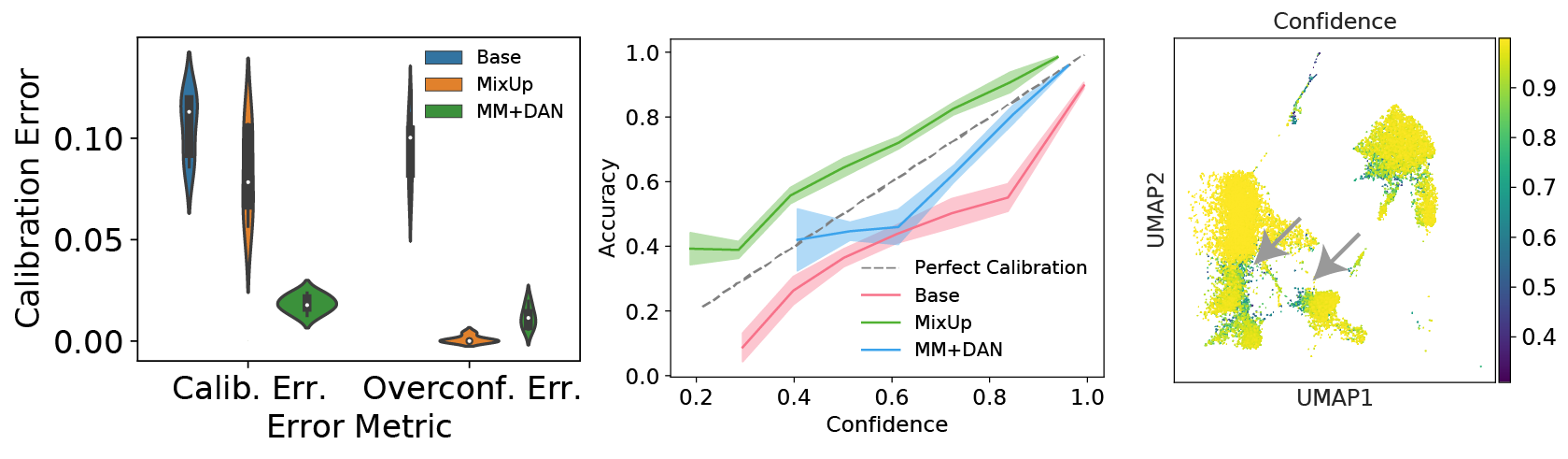
scNym models are well-calibrated and interpretable
scNym confidence scores are well-calibrated
Many machine learning models can provide a metric of classification “confidence,” but these scores do not always correspond to the actual likelihood of prediction accuracy. Models where the confidences scores match the probability of accurate predictions are considered to be well-calibrated. We found that scNym models trained with MixMatch and domain adversaries were better calibrated than standard neural networks, producing useful confidence scores that highlighted incorrect predictions.
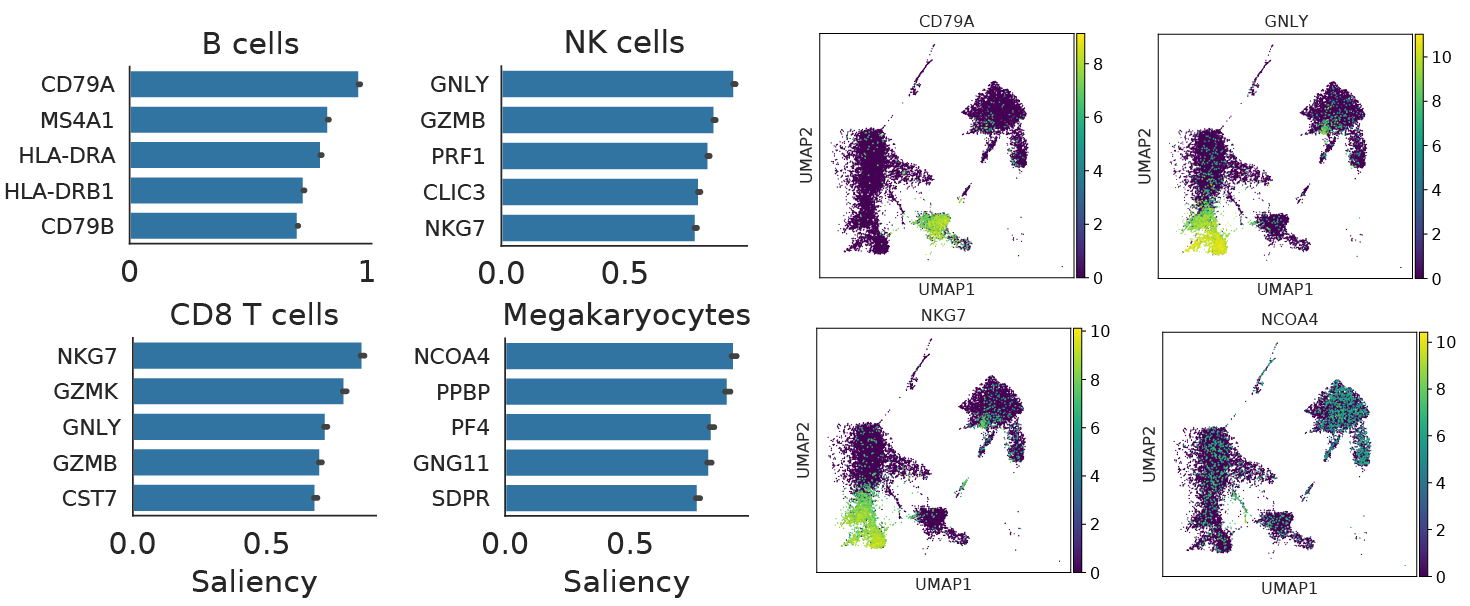
scNym models are interpretable
Neural networks are often described as “black-boxes,” but modern machine learning tools provide a number of methods to interpret model decisions. scNym models can be interpreted using backpropagation-based saliency methods. In short, we compute a gradient on a class score with respect to an input cell profile, asking the question: “Which changes to this profile would make the model more confident about this class?”
We found that these saliency techniques recover many known cell type markers, providing confidence that scNym models learn biologically meaningful relationships between gene expression and cell types. We also found that saliency tools can reveal genes that drive incorrect classification decisions, helping domain experts manually review problematic cases.