Heteromotility
- 2 mins
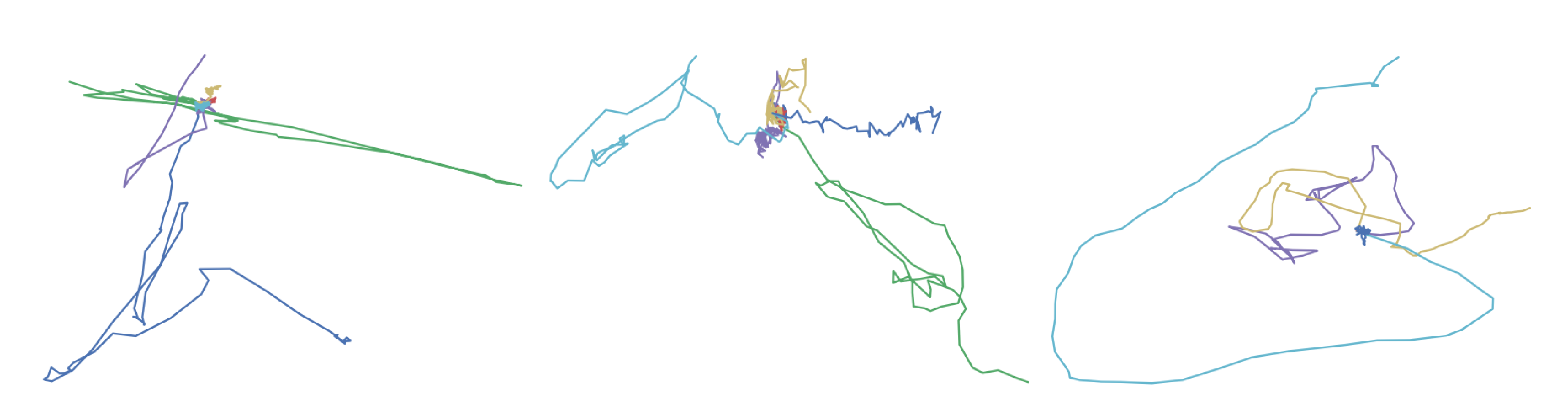
Heteromotility is a tool for analyzing cell motility in a quantitative manner. Heteromotility takes timelapse imaging data as input and calculates various ‘motility features’ that can be used to generate a ‘motility fingerprint’ for a given cell. The tool contains basic image segmentation and cell tracking components, but can also be used to analyze cell trajectories derived from another software tool. By analyzing more features of cell motility than most common cell tracking methods, Heteromotility may be able to identify novel heterogenous motility phenotypes.
Heteromotility also contains a suite of tools to quantify and visualize cell state spaces, and dynamic state transitions within the state space. While these tools were developed for use with Heteromotility features, they may be applied to any arbitrary time-series feature set.
We have a recent paper in PLoS Computational Biology utilizing Heteromotility analysis to quantify dynamic cell state transitions in muscle stem cells and a cancer cell model. Check it out and let me know what you think!
Heteromotility Features
(1) Extraction of 70+ motion features, considering diverse aspects of motility, such as:
- Displacement statistics
- Turning direction and magnitude features
- Features of the cell path in the Cartesian plane
- Comparators to canonical models of motion
- Descriptors of long-term dependence
(2) Simulation of canonical models of motion, including:
- Random walks
- Levy flights
- Fractal Brownian motion
(3) Visualization of cell paths and motility state spaces
(4) Cell state transition quantification tools
(5) Course grained probability flux analysis
Workflow & Usage
Heteromotility functions as a simple command line tool.
Heteromotility can be easily installed as a command line tool from the Python package index (PyPI) using pip.
$ pip install heteromotility
Once installed, Heteromotility is called as a command line utility on CSV files containing N rows of x or y coordinates T units long.
$ heteromotility input_dir output_dir --trackX tracksX.csv --trackY tracksY.csv
The resulting features are saved in output_dir as motility_statistics.csv.
Detailed usage information is available on the Heteromotility GitHub page.
Source code
Help or Suggestions
Please write to me if you have any difficulty or suggestions for improvements!
Cell Motility Video Gallery
A gallery of cell motility videos is available on the Laboratory of Cell Geometry website!
