Rejuvenation By Reprogramming
- 6 mins- Paper: https://doi.org/10.1016/j.cels.2022.05.002
- Paper PDF: PDF Download
- Supplement PDF: PDF Download
- Research Website: reprog.research.calicolabs.com
Mammalian aging dramatically remodels gene expression in diverse cell identities, as revealed by aging cell cartography studies (Calico Murine Aging Cell Atlas, Tabula Muris Senis). Germline ontogeny is the only process known to reverse features of aging in individual cells, such that adult cells can give rise to young animals (Gurdon 1963). Reprogramming cell identity to a pluripotent state the canonical pluripotency transcription factors (Yamanaka factors Sox2, Oct4, Klf4, Myc) has also been reported to erase many features of aging (Mertens et. al. 2015).
Recent reports have suggested that even short, transient activation of the Yamanaka factors is sufficient to reverse some aspects of cellular aging (Ocampo et. al. 2016, Sarkar et. al. 2020, Lu et. al. 2020, Gill et. al.). These exciting results prompt several questions: What features of aging are reversed? Does partial reprogramming exert similar effects across different cell types? Which aspects of the pluripotency program are required for rejuvenation?
Here, we interrogated these questions by mapping trajectories of partial reprogramming in multiple cell types using single cell genomics. We further measured the effect of partial reprogramming with all possible combinations of the Yamanaka factor set using pooled screening approaches. Inspired by limb regeneration in amphibians, we also explored whether partial multipotent reprogramming could restore youthful expression in myogenic cells.
Partial reprogramming restores youthful expression and suppresses cell identity
We performed partial reprogramming with SOKM in young and aged adipogenic and mesenchymal stem cells. By measuring gene expression across single cells, we captured cells in diverse states across the trajectory of partial reprogramming.
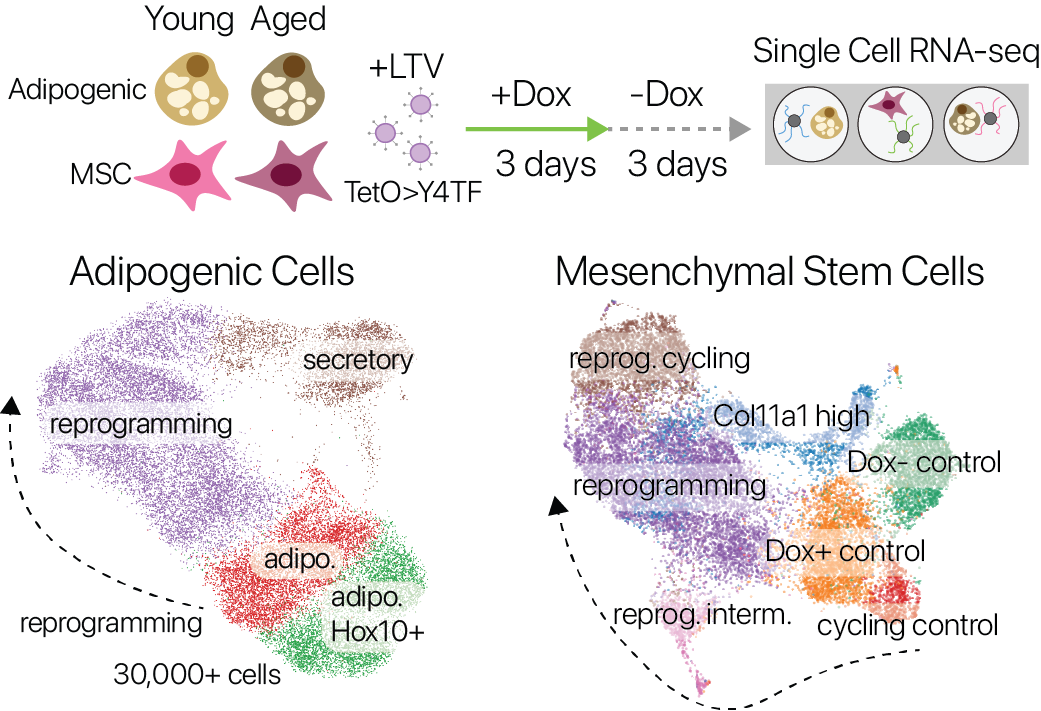
Single cell expression profiles in both adipogenic cells and MSCs revealed a continuous trajectory of cell states induced by partial reprogramming. We also profiled control cells that were not reprogrammed, allowing us to compare the effects of aging and reprogramming in a common measurement space.
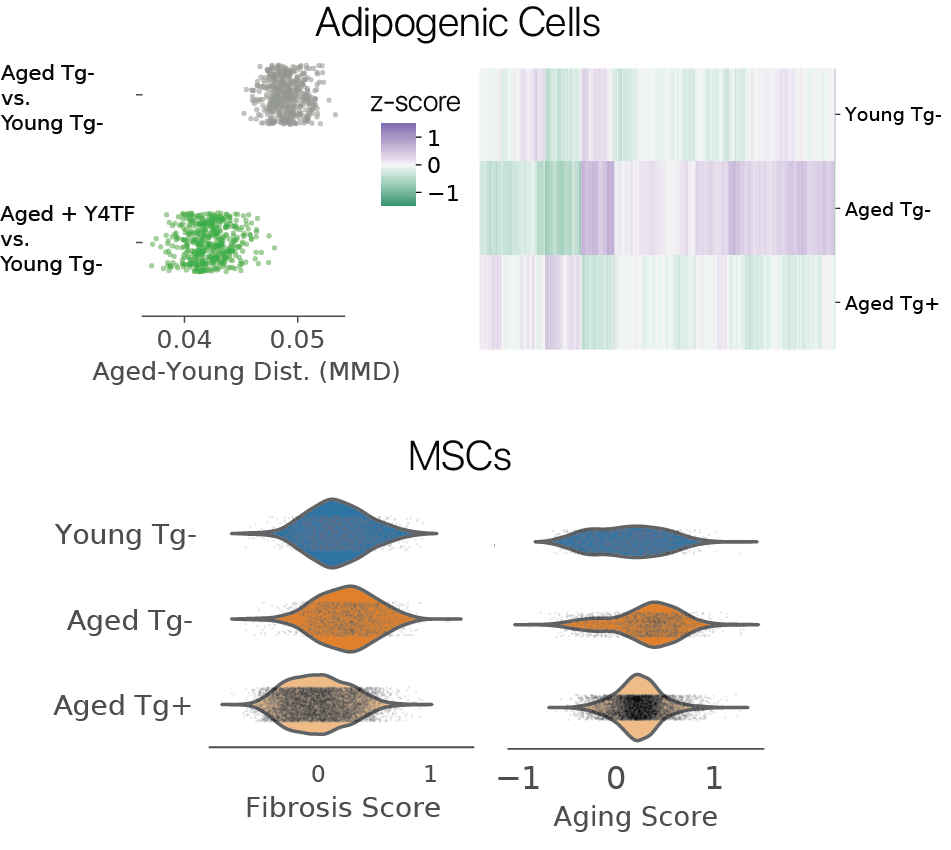
We first wondered if partial reprogramming reversed some features of aging. To investigate, we used maximum mean discrepancy (MMD) comparisons between young and aged cells before and after treatment, considering features across the transcriptome. Remarkably, we found that adipogenic cells were more similar to young controls after treatment, with youthful expression levels restored in thousands of genes. In MSCs, we found that fibrotic gene sets and an aging signature derived from bulk RNA-seq were similarly reduced.
Somatic cell identities are transiently suppressed by partial reprogramming
Reprogramming induced unique cell states, unseen in control conditions in both cell types. These unique states suggested to us that reprogramming might be suppressing somatic cell identity programs, despite some prior reports to the contrary. We performed pseudotime analysis to map each cell to a continuous coordinate system spanning the length of the reprogramming trajectories we observed.
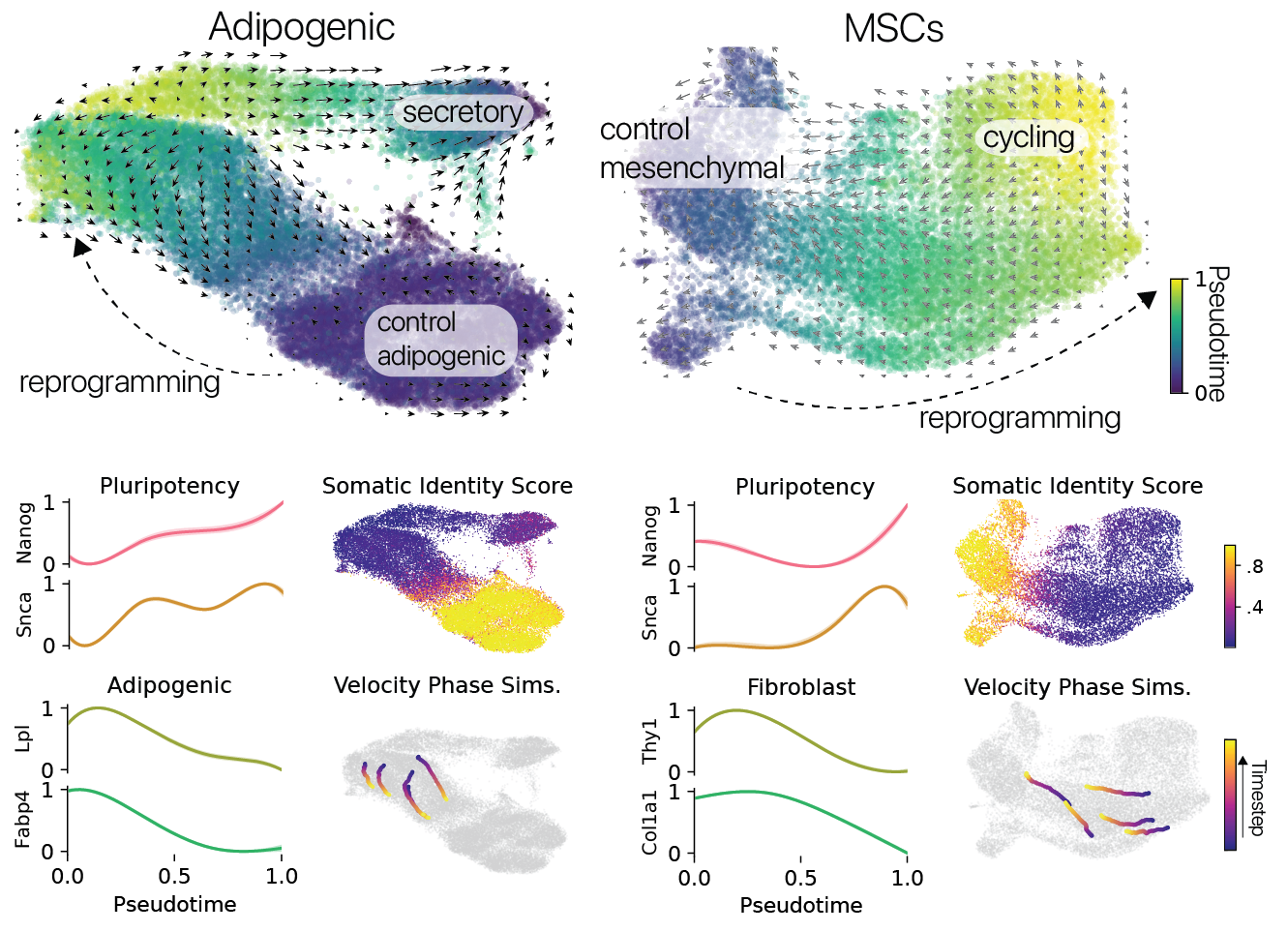
We found that somatic cell identity programs were suppressed and pluripotency identity programs were activated in the most reprogrammed cells along these trajectories. In particular, we observed activation of the Nanog transcription factor, previously reported to be a gate-keeper to the induction of full pluripotency.
Pluripotent cells are characteristically neoplastic, forming teratomas in vivo. Our observation that Nanog is activated in a subset of partially reprogrammed cells suggests that even transient activation of pluripotency programs poses a neoplastic risk. Given that we observed only a small Nanog+ cell population, it seems likely that previous reports using bulk measurements were not able to detect this rare cell state.
We next wondered if partially reprogrammed cells would re-acquire their original somatic identities, as suggested by MEF to iPSC reprogramming systems (Samavarchi-Tehrani et. al. 2010).
We turned to RNA velocity analysis to infer changes in cell state and found that most reprogrammed cells in both populations were re-acquiring their original somatic identities.
Pluripotency submodules are sufficient to restore youthful expression
Are all four Yamanaka factors required to restore youthful expression? Are there any sufficient subsets?
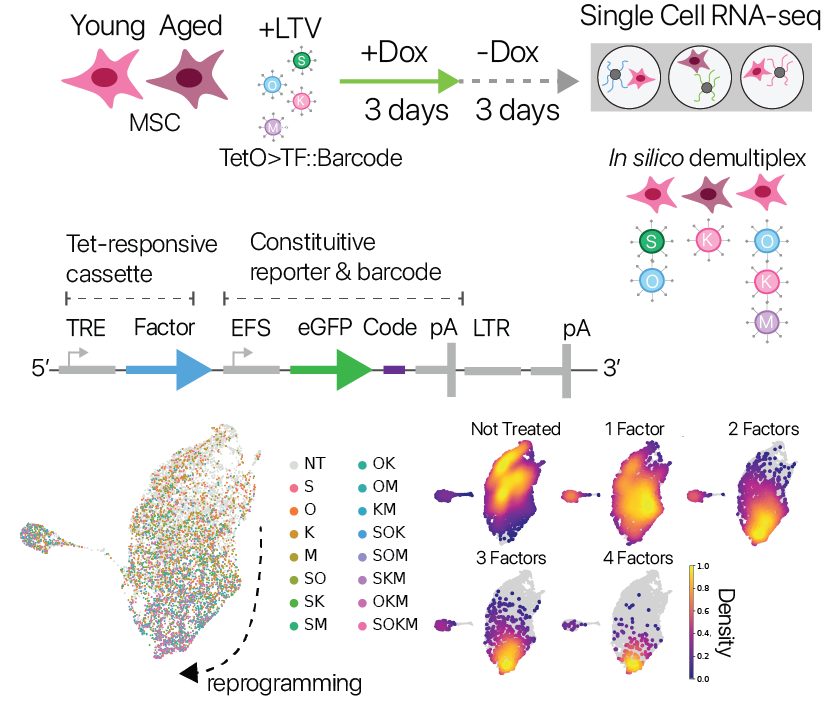
We next wondered if alternative reprogramming strategies could also restore youthful expression. The neoplastic risk posed by oncogenes in the Yamanaka Factor set (Klf4, Myc) motivates a search for alternative approaches. We also wondered if the suppression of cell identity we observed was intimately connected to rejuvenation, or if these two phenomena could be decoupled.
To investigate these questions, we developed a screening system that allowed us to perform partial reprogramming interventions in a pooled format with single cell RNA-seq as a read-out. Our approach was inspired by the CellTag lineage-tracing system (Biddy et. al. 2018), taking advantage of expressed barcodes in the 3’ UTR of a constituitive reporter. We used this system to test partial reprogramming in young and aged MSCs with all possible combinations of the Yamanaka factors.
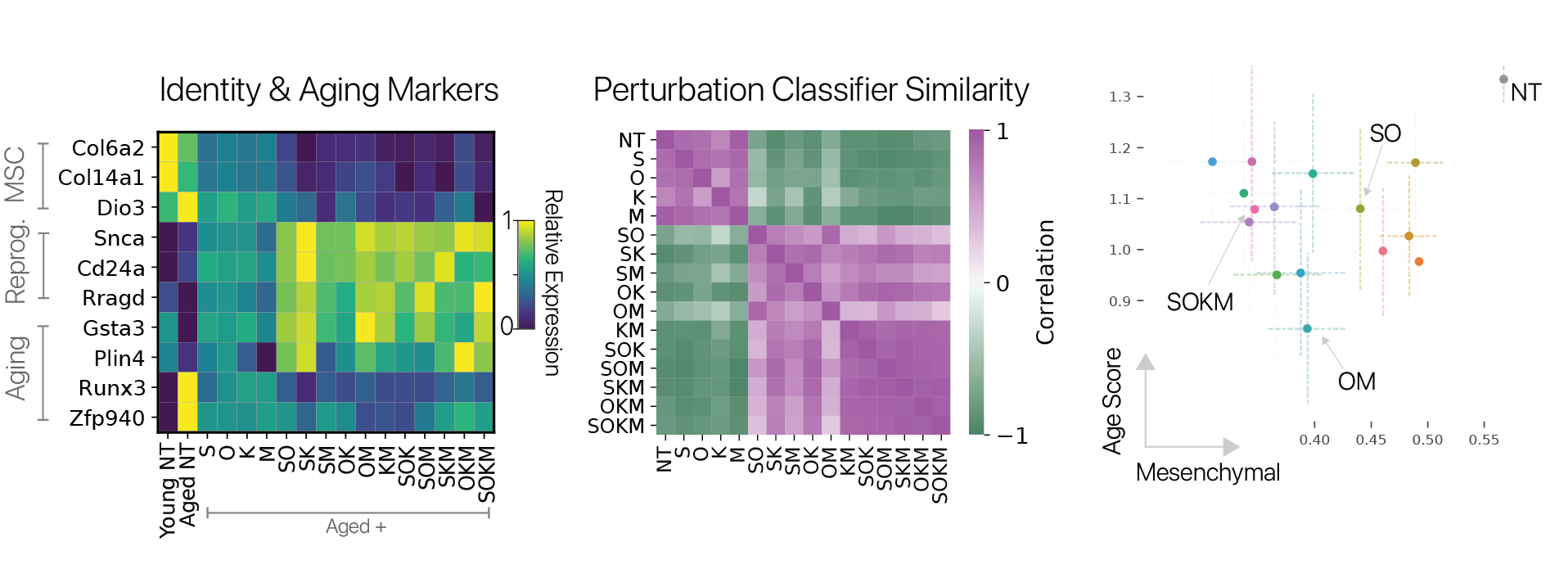
We found that the transcriptional effects of partial reprogramming scaled with the number of unique factors delivered, consistent with known biology for the Yamanaka factors. To determine which combinations had unique effects, we trained a cell identity classification model (scNym) to discriminate different combinations based on transcriptional profiles. We found that effects from combinations of three factors were highly similar to the full Yamanaka factor set, suggesting no single factor is required rejuvenation.
Rejuvenation and identity suppression are not closely entangled
We also scored the expression of an aging gene signature and derived mesenchymal cell identity program scores using a cell classifier trained on a mouse cell atlas (Tabula Muris). We found that almost all combinations significantly reduced the expression of the aging signature, and all significantly suppressed mesenchymal identity. However, the degree of rejuvenation and identity suppression were not significantly correlated, suggesting these effects can be decoupled. The results of our screen suggest that the activation of the full pluripotency program is not required to suppress some features of aging.
Multipotent reprogramming interventions restore myogenic gene expression
Can partial multipotent reprogramming reverse features of aging?
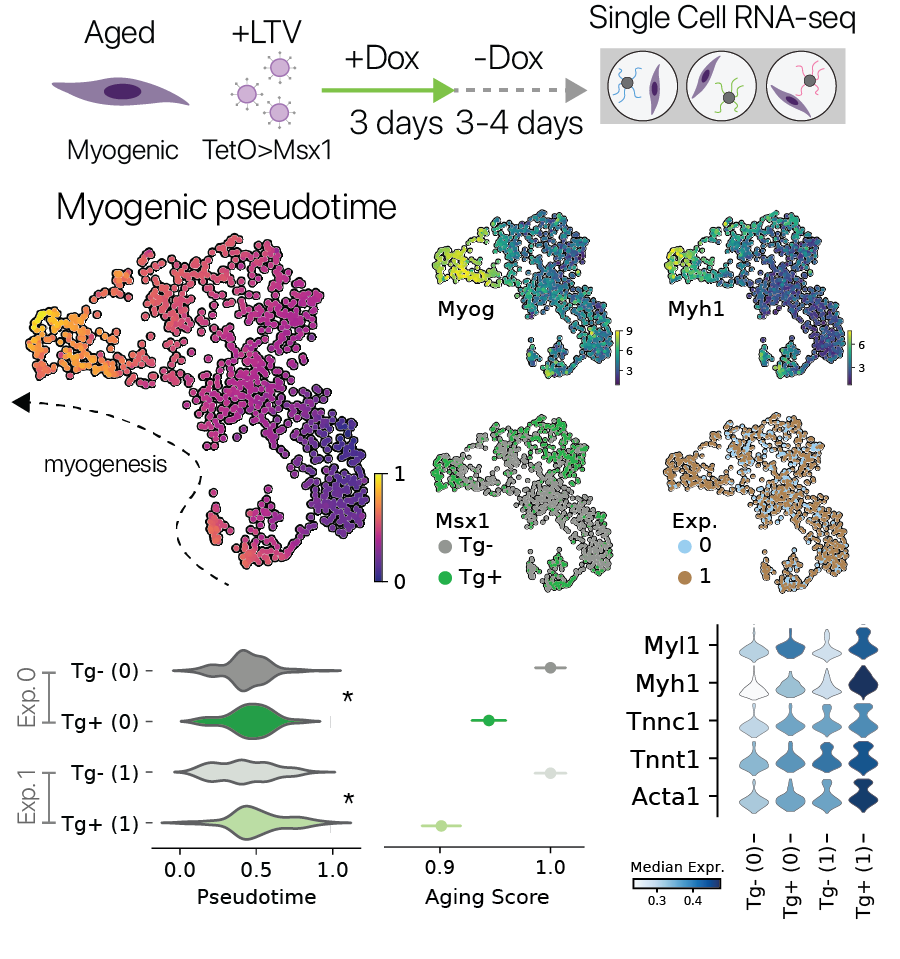
Urodele amphibians have the remarkable ability to regenerate limbs through an endogeneous dedifferentiation process. One key player in this process is the mesodermal transcription factor Msx1. Previous work has shown that Msx1 is sufficient to dedifferentiate synctial myotubes back into proliferating mononuclear progenitor cells, without inducing pluripotency.
We wondered if transient activation of this multipotency factor might also reverse features of aging in myogenic cells, similar to the Yamanaka factors (Sarkar et. al. 2020). We performed a pulse/chase of Msx1 followed by single cell RNA-seq in aged myogenic cells, similar to our other experiments. It has been reported that myogenic differentiation is impaired in aged myogenic cells, and here we found that transient Msx1 treatment improved myogenic gene expression in two independent experiments. This result suggests that transient activation of progenitor factors outside the core pluripotency program may also restore youthful gene expression, similar to the canonical Yamanaka factors.
